A global DNA study reveals a hidden threat in diabetic foot infections
A worldwide DNA analysis reveals that diabetic foot infections are fueled by many different strains of E. coli, not a single culprit.
- Date:
- January 20, 2026
- Source:
- King's College London
- Summary:
- Scientists have uncovered new clues about why diabetic foot infections can become so severe and difficult to treat. By analyzing the DNA of E. coli bacteria taken from infected wounds around the world, researchers found an unexpected level of diversity, with many strains carrying genes linked to antibiotic resistance and aggressive disease. Rather than a single dangerous strain, multiple types of E. coli appear able to thrive in diabetic foot ulcers, helping explain why infections can worsen quickly and sometimes lead to amputation.
- Share:
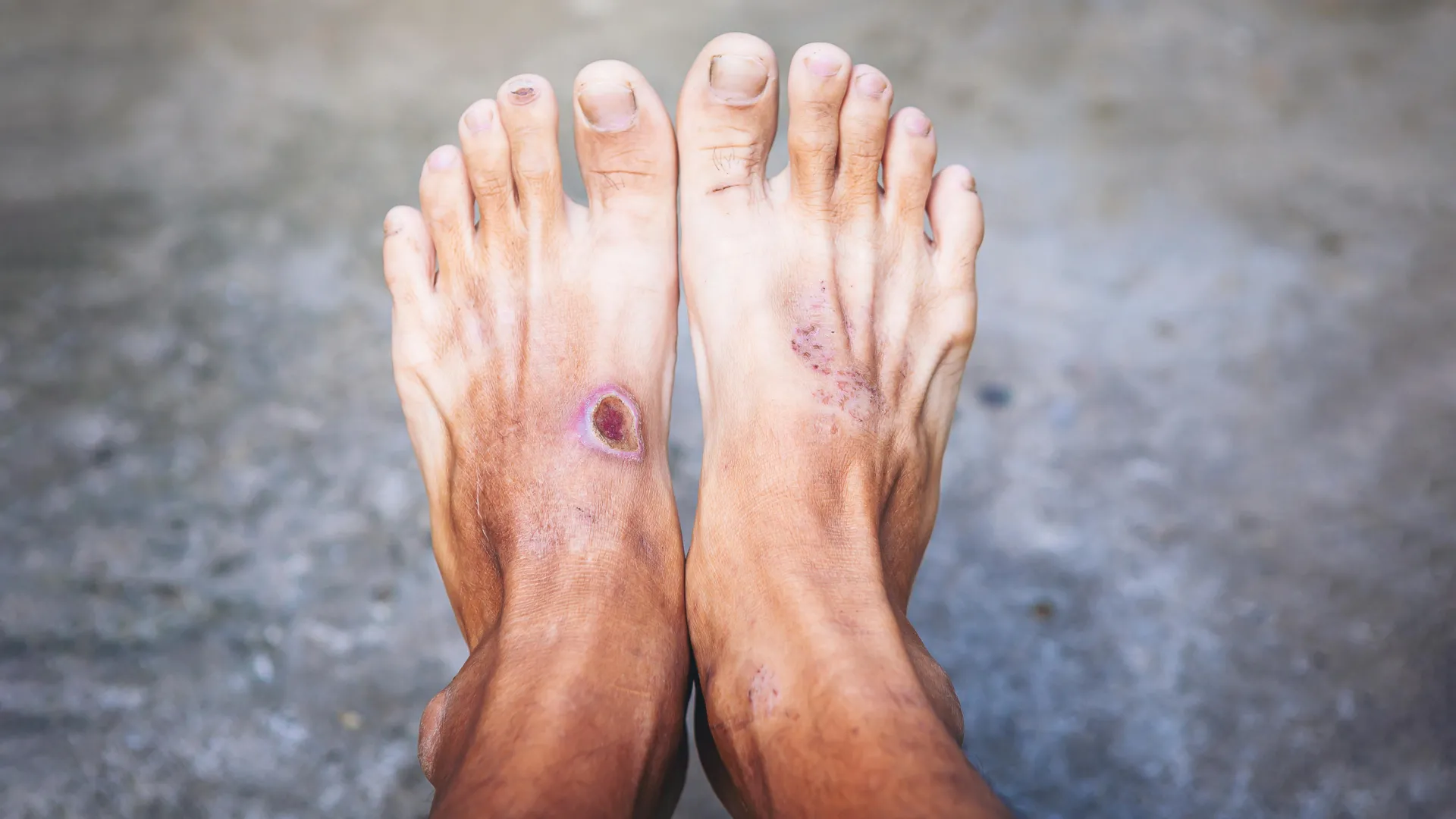
A new study led by King's College London, in partnership with the University of Westminster, has revealed important details about the E. coli bacteria linked to diabetic foot infections. The research focuses on how varied these bacterial strains are and what makes some infections especially severe.
The study, published in Microbiology Spectrum, is the first to offer a large-scale genomic analysis of E. coli taken directly from diabetic foot ulcers across multiple regions of the world. By examining the bacteria at the DNA level, researchers gained insight into why certain infections are difficult to treat and why they can sometimes become life-threatening.
Why Diabetic Foot Infections Are So Dangerous
Diabetic foot infections are among the most serious complications of diabetes and are a leading cause of lower-limb amputation globally. Doctors have long known that these chronic wounds are complex, but there has been limited understanding of the exact microbes involved. This gap in knowledge has been especially noticeable for E. coli, a bacterium that frequently appears in samples from infected wounds but has not been well studied in this context.
Analyzing E. coli From Around the World
To better understand the problem, the research team analyzed complete genome sequences from 42 E. coli strains collected from infected diabetic foot ulcers. The samples came from patients in Nigeria, the UK, Ghana, Sweden, Malaysia, China, South Korea, Brazil, India and the USA. Sequencing the full DNA of each strain allowed scientists to look for global patterns in how E. coli behaves in diabetic foot disease.
This method made it possible to compare genetic differences between strains, detect genes associated with antibiotic resistance, and identify biological traits linked to more severe disease.
No Single Strain Behind These Infections
The genomic results revealed striking diversity among the E. coli strains. The bacteria belonged to many different genetic groups and carried a broad range of genes related to disease and resistance to antibiotics. These findings show that diabetic foot infections are not caused by one specific type of E. coli. Instead, multiple unrelated lineages have independently adapted to survive in the diabetic foot environment.
By examining how the strains are related and which resistance mechanisms and virulence traits (the features or tools that make a microbe more harmful) they possess, the study helps explain why some infections are hard to control or can worsen quickly.
Antibiotic Resistance Raises Concern
One notable finding was that about 8 per cent of the E. coli strains were classified as multidrug-resistant or extensively drug-resistant. This means they can resist several antibiotics or nearly all available treatment options, making infections much more challenging to manage.
Dr. Vincenzo Torraca, Lecturer in Infectious Disease at King's College London and senior author of the study, said: "Understanding these bacteria at a genomic level is a crucial step towards improving diagnosis and enabling more targeted treatments for people with diabetes. By identifying which E. coli strains are most common and which antibiotics they are likely to resist, clinicians can choose therapies that are more likely to work, helping to reduce prolonged infection, hospitalization, and the risk of amputation."
Victor Ajumobi, a second-year PhD student at King's College London and the University of Westminster and first author of the paper, emphasized the broader impact of the findings. "This information will be particularly valuable in low-resource settings, where E. coli infections of diabetic foot ulcers are more common and where rapid diagnostic tools for antimicrobial resistance are not always readily available," he said.
What Comes Next
The researchers plan to continue exploring how specific virulence factors identified in the study influence disease progression. Many of the bacterial samples carry genes that allow E. coli to attach to human tissue or avoid the immune system. Studying how these traits function within diabetic foot wounds could uncover new treatment targets and support the development of more effective therapies.
Story Source:
Materials provided by King's College London. Note: Content may be edited for style and length.
Journal Reference:
- Victor Ajumobi, Zaid Tahir, Polly Hayes, Adele McCormick, Vincenzo Torraca. Population structure, antimicrobial resistance, and virulence factors of diabetic foot-associated Escherichia coli. Microbiology Spectrum, 2026; DOI: 10.1128/spectrum.02837-25
Cite This Page: